Table of Contents
About the Authors
| Whasil Lee University of Rochester, Biomedical Engineering, Pharmacology and Physiology, Center for Musculoskeletal Research |
| Moriana M. Garcia Science & Engineering Outreach Librarian University of Rochester, River Campus Libraries |
About the Tools and Class
| Title | Teaching Students to Explore Protein Structures Using the Visual Molecular Dynamics (VMD) program and 3D Printing. |
| Tool 1 | Visual Molecular Dynamics (VMD) |
| Tool 1 description | VMD is an open-source program designed for modeling, visualization and analysis of biological molecules such as proteins, nucleic acids, etc. It is also a tool to view and analyze the results of molecular dynamic simulations. Using VMD students can open files from the Protein Data Bank (PDB) containing the structure of different biomolecules and analyze them. This tool allows students to connect a protein’s sequence and folding domains with the interaction of specific amino acid residues and the protein’s function; also, to relate disease-associated mutations with protein misfolding and their potential abnormal-functions. |
| Tool 2 | 3D printer at the Rettner Fabrication Studio in Rettner Hall (Mr. James Alkins). |
| Tool 2 description | 3D printer services at the Rettner Fabrication Studio were used to print the 3D structure of selected biomolecules using durable materials. The key advantage of 3D printing is the ability to reproduce very complex protein structures with extreme precision at scale by reading STL files derived from PDB structures using the VMD program. 3D visualization increases student’s comprehension of the link between a biomolecule structure and its functional role |
| Class/Target Level | First year graduate students and motivated undergraduate students |
| Course Title | BME 468 / PHP 468 – Introduction to Structure and Analysis of Biomolecules (Spring 2019) |
| Background information about the class | A series of lectures in BME468 introduced students to the principles of protein & DNA structure and function, fundamental principles of protein folding, DNA structures, the spatial and conceptual relationships of biomolecules, role of mutations, the structure of biomolecules in relation to their functions and dysfunctions – which may be linked directly to human diseases. To visualize and analyze proteins structures, students learned the VMD program. |
| Lesson time | Four classes, each 75 minutes (among 14 classes of a whole semester) |
| Number of students | 10 |
| Learning outcomes | Students completing these four sessions were able to: (1) Know how to visualize and manipulate protein structures from the Protein Data Bank (PDB) using the VMD software. (2) Analyze protein domains, secondary structures and sequences. (3) 3D print PDB files with scaling factors. |
Lesson Plan
Opening
Before the VMD sessions, students learned the structure and function of biomolecules, and how mutations can change a protein conformation and affect its function. Also, they were introduced to NCBI databases, PubMed, and BLAST sequence alignment resources by a librarian.
Body
During the VMD sessions, students learned the basic steps of working with the software as described below:
Module 1
Install the VMD program and open PDB files of proteins of interest.
In this module, students downloaded and installed the VMD program (https://www.ks.uiuc.edu/Research/vmd/) in desktop or laptop computers. Following the installation, students learned the basic VMD commands. To practice, all students downloaded and opened the same protein – Piezo1 PDB (http://www.rcsb.org/structure/6B3R). After that, students selected their proteins of interest, downloaded the PDB file from the PDB Bank, and visualized their 3D structure using VMD.
Module 2
Analyze secondary and tertiary structure of proteins, and protein sequence.
In this module, students learned the ‘Graphical Representations’ options in the VMD program. Students selected different styles and colors to display secondary structures and folding domains. Students also learned the ‘Sequence Viewer Extension’ options in VMD, an option that allows students to find the location of selected amino acids easily.
Module 3
Detect bonding interactions, save images.
First, students generated a PSF file from their protein of interest using the ‘Automatic PSF Builder’ in VMD. A PSF file stores the structural information of proteins, such as various types of bonding interactions. After loading both PSF and PDB files together in VMD, students were able to visualize hydrogen bonds and ionic bonds. Also, selecting the representation in ‘charge’, students were able to visualize the hydrophobic patches or hydrophilic regions. After these steps, students learned how to save their visualization and how to render publication-quality images of molecules. Based on information gathered from the literature related to associated functional domains or disease associated mutation sites, students found the regions of interest in their protein structures and saved the images (Fig. 1), or a series of images to generate a movie (Suppl. Mov. 1).
Module 4
3D-print the protein of interest.
In this module, students learned how to export PDB files into STL files in order to 3D-print their chosen molecule. Students measured the size of their proteins and calculated a scaling factor to make an approximately 5cm x 5cm x 5cm print-out. With Mr. Alkins’ guidance, students uploaded their STL files and submitted them for printing.
Closing
After the basic VMD training and some practice visualizing their proteins of interest, each student prepared an oral presentation describing their protein ‘s conformation, folding domains, sequence homology among different species and mutation-associated human diseases. During the presentation, students also used the protein 3D-printout to point relevant folding domains and the location of mutations on the molecule.
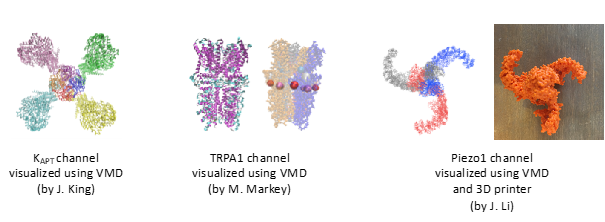
Images J. King, M. Markey, J. Li, with permission
Findings/assessment
The instructor and peer students evaluated each students’ presentation using a rubric evaluating Content, Effort, Originality and Visualization, with the last category being the one linked to the use of VMD.
Accommodation/differentiation
Although no accommodation or differentiation was required during this instance of the course, students with visual impairment would benefit from the use of 3D printed structures, which allows them to understand the spatial representation of their protein through active tactile perception and haptic exploration.
Reflection
Reflection from instructor
The use of VMD and 3D printing allowed the instructor to teach students an in-depth analysis of atomic-scaled biomolecules. Via hands-on experience, students learned how to visualize and analyze biomolecules step by step, from learning the basic stabilizing atomic interactions between atoms to understanding the folding domains and conformational changes of proteins. In sum, the inclusion of these digital tools in the class environment boosted students’ learning by showing them how three-dimensional changes in the structure of biomolecules allow them to function efficiently.
Preparation time/materials
The VMD tutorial is adapted from https://www.ks.uiuc.edu/Training/Tutorials/vmd/tutorial-html/.
Benefits and challenges of the tool
Benefits: Gaining mastery of these tools empowers students to apply the acquired knowledge to future research projects.
Challenges: Some of the challenges of the tool are related to PDB files with incomplete versions, students had to open several files to find one they could 3D-print. In addition, some STL files are too big (GB), forcing students to minimize the file size by taking the visualization as a Surface option with less detail.
Ease of use/ranking
Beginner to advanced
Reflection from students
Student comment from the ‘Course Evaluation Results’ of BME468 Spring 2019.
Q: What are the major strengths of this course?
A: The use of the VMD program and the critical thinking required in the later half of the class. I believe the group paper discussion and the individual presentations were particularly strong parts of the course which allowed students to communicate with others and learn what they might not know/think further about the topics of research.